Current Projects
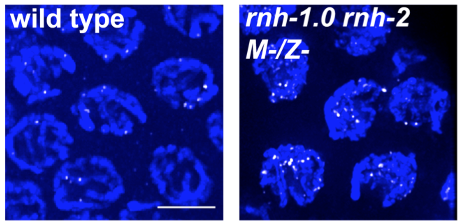
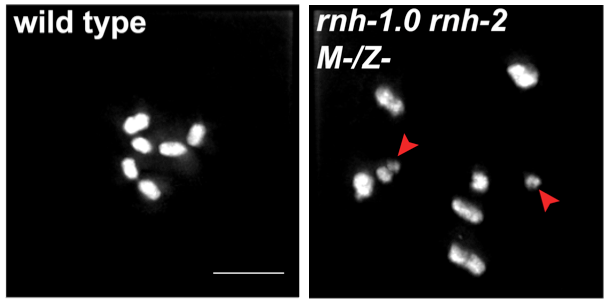
1) R-loops in meiosis. R-loops are nucleic acid structures composed of DNA-RNA hybrid and displaced ssDNA. It is established that when R-loops aberrantly accumulate they create replication-stress that leads to DSB formation. We have shown that R-loop accumulation in germ-cells also results in DSBs, but these fail to properly repair, avoiding repair via both homologous recombination and end joining pathways (Hicks et al 2022). Moreover, nuclei harboring this irreparable DSBs, do not properly activate cell cycle arrest and apoptosis checkpoints, leading to persistence of irreparable DSBs in oocytes. We are currently conducting studies to identify how R-loops-generated DSBs avoid repair and detection and how they influence other events in meiosis and embryogenesis.
DSB accumulation (top, RAD-51) and DNA fragmentation (bottom, DAPI) as result of R-loop accumulation in the germline
2) Meiotic DSBs are form continuously and are repaired by different sub-pathways. In collaboration with the Silva lab (https://biology.med.muni.cz/en/research/nicola-silva-group/about) we discovered that meiotic DSBs form continually from meiotic entry to late pachytene transition (Hicks et al 2022). Although early and late DSBs are both repaired by homologous recombination, they are targeted to different outcomes within this pathway: early DSBs non-crossover and late DSBs to crossovers. We are currently investigating the mechanism regulation of these two faits in the context of early and late DSBs.
A working model for early and late meiotic DSBs 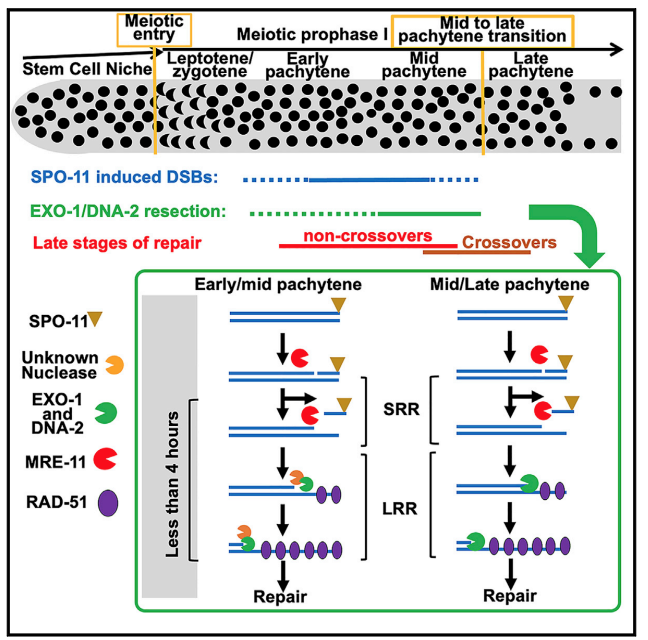
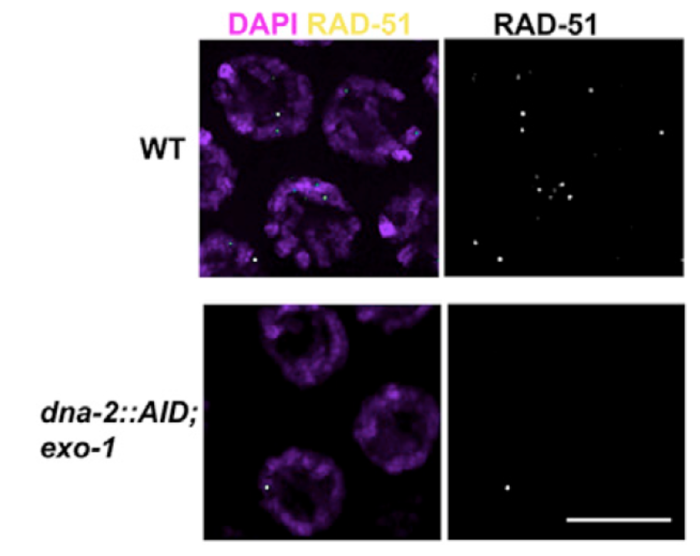
3) Early events in meiotic double strand breaks (DSB) processing in the germline and DSB repair pathway choice. In the germline DSBs are repaired by homologous recombination, a primarily error free repair pathway. A key event in DSb repair is resection, the processing of the ends of the DSBs to ssDNA. We discovered that in the C. elegans germline these ssDNA is substrate for multiple RPA complexes (Hefel et al 2021). The formation of ssDNA is orchestrated by several proteins divided roughly into short and long range resection based on the extent of resection executed by these mechanisms. Our lab identified a separation of function allele of MRE-11, which abrogates the resection activity of this protein without affecting its role in meiotic DSB formation (Yin and Smolikove 2013). In this mutants DSB short range resection is abrogated and repair now occurs by an error prone repair pathway- classical non homologous end joining (cNHEJ). We use this mutant as well as MRE-11 screening approaches as a tool for identifying novel meiotic gene involved in DSB repair in C. elegans and how repair pathway choice between HR and NHEJ is regulated (Yin et. al. 2016, Reichman et. al. 2018). We recently discovered that DNA-2 and EXO-1 play redundant roles in meiotic long-range resection and that their activity is only partially required for early DSBs (Hicks et al 2022).
Removing EXO-1 and DNA-2 abrogates resection in meiosis
4) Repair of complex DNA damage in meiosis. We have developed a method to detect recruitment of DSB repair proteins to sites of DSBs in live, intact, worms (Koury et al 2017, Harrell et al 2018). We use this method to study how complex DNA damage is repaired in the germline. We identify the recruitment kinetics proteins involved in HR and NHEJ in different regions of the germline and in mitotic vs. meiotic nuclei of the worm. We have shown that complex DNA damage involves both the HR and NHEJ repair pathways (Koury et al 2017, Harrell et al. 2021) and currently investigate other unique aspects of this repair.
We welcome collaborations with our lab using this method
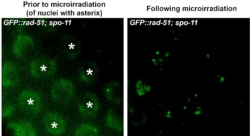
Forming DSBs in live worms by laser and following protein recruitment to DSBs in vivo in intact animals
Other projects
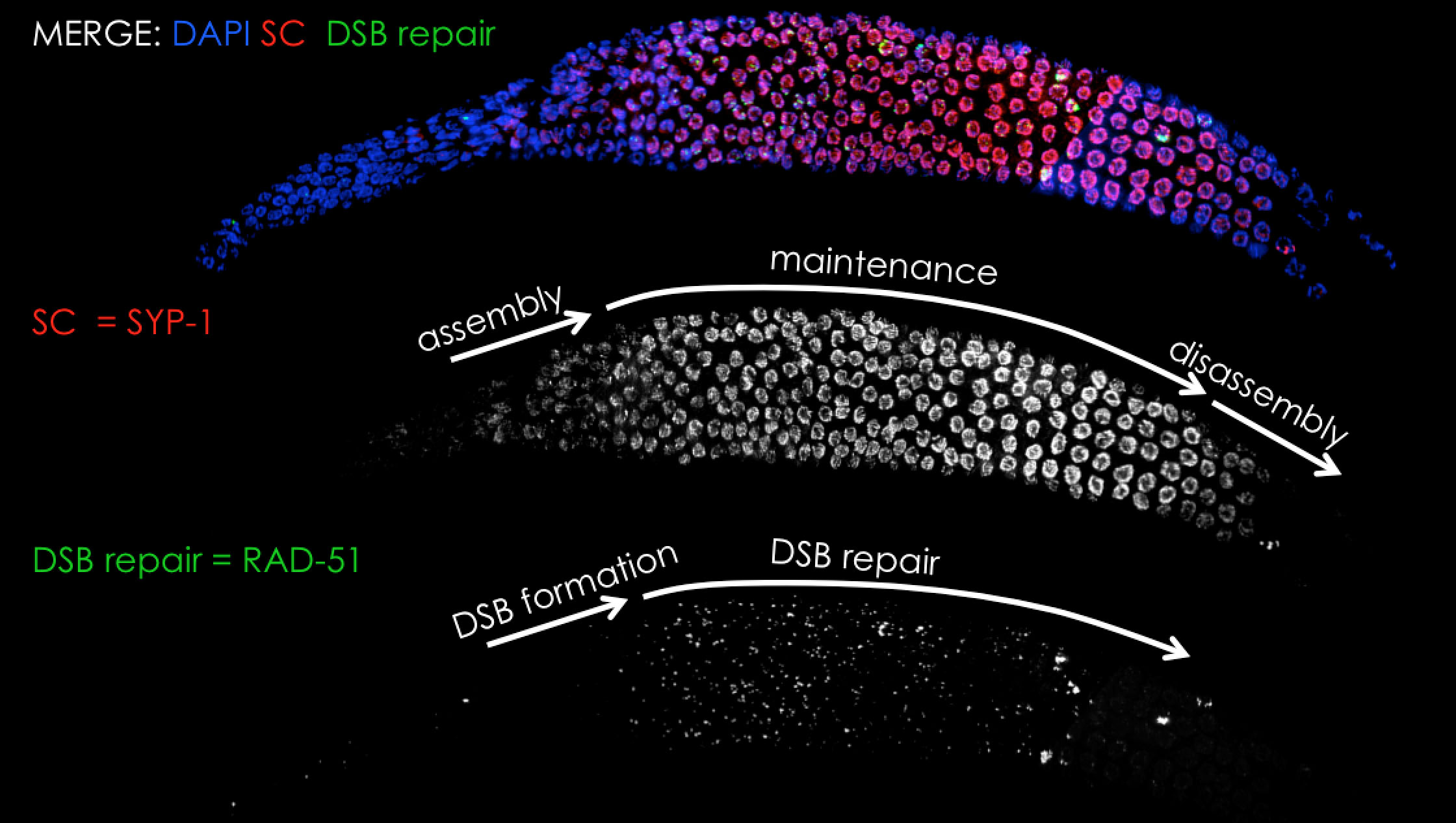 We have long time experience in analysis of the synaptonemal complex in C. elegans meiosis using cell-biology and genetic based methods (see publications page). Although we do not have currently an active project on this topic we are happy to share our experience and collaborate on this topic.
We have long time experience in analysis of the synaptonemal complex in C. elegans meiosis using cell-biology and genetic based methods (see publications page). Although we do not have currently an active project on this topic we are happy to share our experience and collaborate on this topic.